{{How to cite NGSmethDB}}
As an example, we will retrieve the methylation levels of single cytosines of the AMH gene in psoas muscle. This gene codes for the anti-Mullerian hormone, a member of the transforming growth factor-beta gene family which mediates male sexual differentiation.
1. Go to NGSmethDB and then click on Data access -> Track hubs -> Single-cytosine methylation -> Human – hg38 -> Genome map at UCSC.
2. Once at UCSC, type AMH on the search text field, then select ‘AMH (Homo sapiens anti-Mullerian hormone (AMH), mRNA. (from RefSeq NM_000479))’ and finally click go.
3. The browser then will show the genome region of chromosome 19 where the AMH gene is located: chr19:2,249,311-2,252,073.
4. Click Tools -> Table Browser.
5. In group choose ‘NGSmethDB: Methylation levels’ and in table choose the sample ‘STL001_psoasMuscle’.
6. Click get output to obtain the result table in your browser (only a few rows and columns of the table are shown):
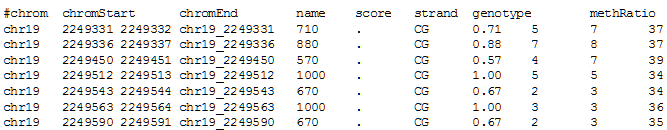
7. Go back with your browser to the Table Browser page and then click on summary/statistics to get a summary of your results:
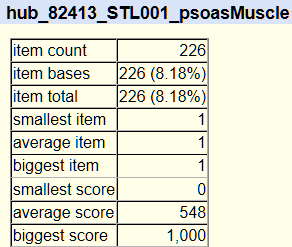
8. If you want to retrieve data for the entire chromosome, go back to the Table Browser page and in region -> position type ‘chr2’ (or whatever else chromosome number you want). To get data for the entire genome, in region click on the button ‘genome’.
9. If you want to save the numerical data to your hard disk, go back to the Table Browser page and then provide a filename in the output file text field.
10. Finally, if you want to send your numerical data to the Galaxy platform for further downstream analyses, go back to the Table Browser page and then tick on Send output to Galaxy.
[[How to cite NGSmethDB]]
- Geisen et al. (2014) Nucleic Acids Res, 42, D53-9. Free Full Text
- Hackenberg et al. (2011) Nucleic Acids Res, 39, D75-9. Free Full Text
[[How to cite NGSmethDB]]